ARG53958
anti-p53 antibody [BP53-12] (FITC)
anti-p53 antibody [BP53-12] (FITC) for Flow cytometry and Human,Primates
Cancer antibody; Cell Biology and Cellular Response antibody; Cell Death antibody; Gene Regulation antibody
Overview
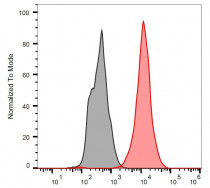
| Product Description | FITC-conjugated Mouse Monoclonal antibody [BP53-12] recognizes p53 |
|---|---|
| Tested Reactivity | Hu, NHuPrm |
| Tested Application | FACS |
| Specificity | The clone BP53-12 recognizes defined epitope (aa 16-25) on human p53, a 50 kDa tumour suppressor found in increased amounts in a wide variety of transformed cells; it is frequently mutated or inactivated in many types of cancer. |
| Host | Mouse |
| Clonality | Monoclonal |
| Clone | BP53-12 |
| Isotype | IgG2a |
| Target Name | p53 |
| Antigen Species | Human |
| Immunogen | Bacterially expressed full-length wild-type p53 |
| Epitope | The epitope of the antibody is around aa 16-25 of human p53 |
| Conjugation | FITC |
| Alternate Names | TRP53; LFS1; Cellular tumor antigen p53; Phosphoprotein p53; Tumor suppressor p53; BCC7; Antigen NY-CO-13; P53 |
Application Instructions
| Application Suggestion |
|
||||
|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. |
Properties
| Form | Liquid |
|---|---|
| Purification Note | The purified antibody is conjugated with Fluorescein isothiocyanate (FITC) under optimum conditions. The reagent is free of unconjugated FITC. |
| Buffer | PBS (pH 7.4) and 15 mM Sodium azide |
| Preservative | 15 mM Sodium azide |
| Concentration | 1 mg/ml |
| Storage Instruction | Aliquot and store in the dark at 2-8°C. Keep protected from prolonged exposure to light. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links | |
|---|---|
| Gene Symbol | TP53 |
| Gene Full Name | tumor protein p53 |
| Background | p53 is a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Mutations in this gene are associated with a variety of human cancers, including hereditary cancers such as Li-Fraumeni syndrome. Alternative splicing of this gene and the use of alternate promoters result in multiple transcript variants and isoforms. Additional isoforms have also been shown to result from the use of alternate translation initiation codons from identical transcript variants (PMIDs: 12032546, 20937277). [provided by RefSeq, Dec 2016] |
| Function | p53 acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. One of the activated genes is an inhibitor of cyclin-dependent kinases. Apoptosis induction seems to be mediated either by stimulation of BAX and FAS antigen expression, or by repression of Bcl-2 expression. Its pro-apoptotic activity is activated via its interaction with PPP1R13B/ASPP1 or TP53BP2/ASPP2 (PubMed:12524540). However, this activity is inhibited when the interaction with PPP1R13B/ASPP1 or TP53BP2/ASPP2 is displaced by PPP1R13L/iASPP (PubMed:12524540). In cooperation with mitochondrial PPIF is involved in activating oxidative stress-induced necrosis; the function is largely independent of transcription. Induces the transcription of long intergenic non-coding RNA p21 (lincRNA-p21) and lincRNA-Mkln1. LincRNA-p21 participates in TP53-dependent transcriptional repression leading to apoptosis and seems to have an effect on cell-cycle regulation. Implicated in Notch signaling cross-over. Prevents CDK7 kinase activity when associated to CAK complex in response to DNA damage, thus stopping cell cycle progression. Isoform 2 enhances the transactivation activity of isoform 1 from some but not all TP53-inducible promoters. Isoform 4 suppresses transactivation activity and impairs growth suppression mediated by isoform 1. Isoform 7 inhibits isoform 1-mediated apoptosis. Regulates the circadian clock by repressing CLOCK-ARNTL/BMAL1-mediated transcriptional activation of PER2 (PubMed:24051492). [UniProt] |
| Highlight | Related products: p53 antibodies; p53 ELISA Kits; p53 Duos / Panels; p53 recombinant proteins; Anti-Mouse IgG secondary antibodies; Related news: Therapeutic strategies against PDAC Senescence Marker Antibody Panel is launched |
| Research Area | Cancer antibody; Cell Biology and Cellular Response antibody; Cell Death antibody; Gene Regulation antibody |
| Calculated MW | 44 kDa |
| PTM | Acetylated. Acetylation of Lys-382 by CREBBP enhances transcriptional activity. Deacetylation of Lys-382 by SIRT1 impairs its ability to induce proapoptotic program and modulate cell senescence. Deacetylation by SIRT2 impairs its ability to induce transcription activation in a AKT-dependent manner. Phosphorylation on Ser residues mediates transcriptional activation. Phosphorylated by HIPK1 (By similarity). Phosphorylation at Ser-9 by HIPK4 increases repression activity on BIRC5 promoter. Phosphorylated on Thr-18 by VRK1. Phosphorylated on Ser-20 by CHEK2 in response to DNA damage, which prevents ubiquitination by MDM2. Phosphorylated on Ser-20 by PLK3 in response to reactive oxygen species (ROS), promoting p53/TP53-mediated apoptosis. Phosphorylated on Thr-55 by TAF1, which promotes MDM2-mediated degradation. Phosphorylated on Ser-33 by CDK7 in a CAK complex in response to DNA damage. Phosphorylated on Ser-46 by HIPK2 upon UV irradiation. Phosphorylation on Ser-46 is required for acetylation by CREBBP. Phosphorylated on Ser-392 following UV but not gamma irradiation. Phosphorylated on Ser-15 upon ultraviolet irradiation; which is enhanced by interaction with BANP. Phosphorylated by NUAK1 at Ser-15 and Ser-392; was intially thought to be mediated by STK11/LKB1 but it was later shown that it is indirect and that STK11/LKB1-dependent phosphorylation is probably mediated by downstream NUAK1 (PubMed:21317932). It is unclear whether AMP directly mediates phosphorylation at Ser-15. Phosphorylated on Thr-18 by isoform 1 and isoform 2 of VRK2. Phosphorylation on Thr-18 by isoform 2 of VRK2 results in a reduction in ubiquitination by MDM2 and an increase in acetylation by EP300. Stabilized by CDK5-mediated phosphorylation in response to genotoxic and oxidative stresses at Ser-15, Ser-33 and Ser-46, leading to accumulation of p53/TP53, particularly in the nucleus, thus inducing the transactivation of p53/TP53 target genes. Phosphorylated by DYRK2 at Ser-46 in response to genotoxic stress. Phosphorylated at Ser-315 and Ser-392 by CDK2 in response to DNA-damage. Dephosphorylated by PP2A-PPP2R5C holoenzyme at Thr-55. SV40 small T antigen inhibits the dephosphorylation by the AC form of PP2A. May be O-glycosylated in the C-terminal basic region. Studied in EB-1 cell line. Ubiquitinated by MDM2 and SYVN1, which leads to proteasomal degradation (PubMed:10722742, PubMed:12810724, PubMed:15340061, PubMed:17170702, PubMed:19880522). Ubiquitinated by RFWD3, which works in cooperation with MDM2 and may catalyze the formation of short polyubiquitin chains on p53/TP53 that are not targeted to the proteasome (PubMed:10722742, PubMed:12810724, PubMed:20173098). Ubiquitinated by MKRN1 at Lys-291 and Lys-292, which leads to proteasomal degradation (PubMed:19536131). Deubiquitinated by USP10, leading to its stabilization (PubMed:20096447). Ubiquitinated by TRIM24, RFFL, RNF34 and RNF125, which leads to proteasomal degradation (PubMed:19556538). Ubiquitination by TOPORS induces degradation (PubMed:19473992). Deubiquitination by USP7, leading to stabilization (PubMed:15053880). Isoform 4 is monoubiquitinated in an MDM2-independent manner (PubMed:15340061). Ubiquitinated by RFWD2, which leads to proteasomal degradation (PubMed:19837670). Ubiquitination and subsequent proteasomal degradation is negatively regulated by CCAR2 (PubMed:25732823). Monomethylated at Lys-372 by SETD7, leading to stabilization and increased transcriptional activation. Monomethylated at Lys-370 by SMYD2, leading to decreased DNA-binding activity and subsequent transcriptional regulation activity. Lys-372 monomethylation prevents interaction with SMYD2 and subsequent monomethylation at Lys-370. Dimethylated at Lys-373 by EHMT1 and EHMT2. Monomethylated at Lys-382 by KMT5A, promoting interaction with L3MBTL1 and leading to repress transcriptional activity. Dimethylation at Lys-370 and Lys-382 diminishes p53 ubiquitination, through stabilizing association with the methyl reader PHF20. Demethylation of dimethylated Lys-370 by KDM1A prevents interaction with TP53BP1 and represses TP53-mediated transcriptional activation. Sumoylated with SUMO1. Sumoylated at Lys-386 by UBC9. [UniProt] |
Images (1) Click the Picture to Zoom In
Clone References