ARG51752
anti-Histone H2A.X phospho (Ser139) antibody
anti-Histone H2A.X phospho (Ser139) antibody for ICC/IF,Western blot and Human
Gene Regulation antibody
Overview
| Product Description | Rabbit Polyclonal antibody recognizes Histone H2A.X phospho (Ser139) |
|---|---|
| Tested Reactivity | Hu |
| Tested Application | ICC/IF, WB |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Target Name | Histone H2A.X |
| Antigen Species | Human |
| Immunogen | Peptide sequence around phosphorylation site of serine 139 (Q-A-S(p)-Q-E) derived from Human Histone H2A.X. |
| Conjugation | Un-conjugated |
| Alternate Names | H2AX; H2a/x; H2A.X; Histone H2AX; H2A/X; Histone H2A.X; gamma H2AX; gamma H2A.X |
Application Instructions
| Application Suggestion |
|
||||||
|---|---|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. |
Properties
| Form | Liquid |
|---|---|
| Purification | Antibodies were produced by immunizing rabbits with KLH-conjugated synthetic phosphopeptide. Antibodies were purified by affinity-chromatography using epitope-specific phosphopeptide. In addition, non-phospho specific antibodies were removed by chromatogramphy using non-phosphopeptide. |
| Buffer | PBS (without Mg2+ and Ca2+, pH 7.4), 150mM NaCl, 0.02% Sodium azide and 50% Glycerol. |
| Preservative | 0.02% Sodium azide |
| Stabilizer | 50% Glycerol |
| Concentration | 1 mg/ml |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links | |
|---|---|
| Gene Symbol | H2AFX |
| Gene Full Name | H2A histone family, member X |
| Background | Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation. |
| Function | Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation. [UniProt] |
| Highlight | Related products: Histone H2A.X antibodies; Anti-Rabbit IgG secondary antibodies; Related news: Senescence Marker Antibody Panel is launched |
| Research Area | Gene Regulation antibody |
| Calculated MW | 15 kDa |
| PTM | Phosphorylated on Ser-140 (to form gamma-H2AX or H2AX139ph) in response to DNA double strand breaks (DSBs) generated by exogenous genotoxic agents and by stalled replication forks, and may also occur during meiotic recombination events and immunoglobulin class switching in lymphocytes. Phosphorylation can extend up to several thousand nucleosomes from the actual site of the DSB and may mark the surrounding chromatin for recruitment of proteins required for DNA damage signaling and repair. Widespread phosphorylation may also serve to amplify the damage signal or aid repair of persistent lesions. Phosphorylation of Ser-140 (H2AX139ph) in response to ionizing radiation is mediated by both ATM and PRKDC while defects in DNA replication induce Ser-140 phosphorylation (H2AX139ph) subsequent to activation of ATR and PRKDC. Dephosphorylation of Ser-140 by PP2A is required for DNA DSB repair. In meiosis, Ser-140 phosphorylation (H2AX139ph) may occur at synaptonemal complexes during leptotene as an ATM-dependent response to the formation of programmed DSBs by SPO11. Ser-140 phosphorylation (H2AX139ph) may subsequently occurs at unsynapsed regions of both autosomes and the XY bivalent during zygotene, downstream of ATR and BRCA1 activation. Ser-140 phosphorylation (H2AX139ph) may also be required for transcriptional repression of unsynapsed chromatin and meiotic sex chromosome inactivation (MSCI), whereby the X and Y chromosomes condense in pachytene to form the heterochromatic XY-body. During immunoglobulin class switch recombination in lymphocytes, Ser-140 phosphorylation (H2AX139ph) may occur at sites of DNA-recombination subsequent to activation of the activation-induced cytidine deaminase AICDA. Phosphorylation at Tyr-143 (H2AXY142ph) by BAZ1B/WSTF determines the relative recruitment of either DNA repair or pro-apoptotic factors. Phosphorylation at Tyr-143 (H2AXY142ph) favors the recruitment of APBB1/FE65 and pro-apoptosis factors such as MAPK8/JNK1, triggering apoptosis. In contrast, dephosphorylation of Tyr-143 by EYA proteins (EYA1, EYA2, EYA3 or EYA4) favors the recruitment of MDC1-containing DNA repair complexes to the tail of phosphorylated Ser-140 (H2AX139ph). Monoubiquitination of Lys-120 (H2AXK119ub) by RING1 and RNF2/RING2 complex gives a specific tag for epigenetic transcriptional repression (By similarity). Following DNA double-strand breaks (DSBs), it is ubiquitinated through 'Lys-63' linkage of ubiquitin moieties by the E2 ligase UBE2N and the E3 ligases RNF8 and RNF168, leading to the recruitment of repair proteins to sites of DNA damage. Ubiquitination at Lys-14 and Lys-16 (H2AK13Ub and H2AK15Ub, respectively) in response to DNA damage is initiated by RNF168 that mediates monoubiquitination at these 2 sites, and 'Lys-63'-linked ubiquitin are then conjugated to monoubiquitin; RNF8 is able to extend 'Lys-63'-linked ubiquitin chains in vitro. H2AK119Ub and ionizing radiation-induced 'Lys-63'-linked ubiquitination (H2AK13Ub and H2AK15Ub) are distinct events. Acetylation at Lys-37 increases in S and G2 phases. This modification has been proposed to play a role in DNA double-strand break repair (By similarity). |
Images (3) Click the Picture to Zoom In
-
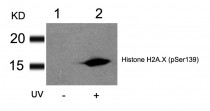
ARG51752 anti-Histone H2A.X phospho (Ser139) antibody WB image
Western blot: Extracts from HT29 cells untreated(lane 1) or treated with UV(lane 2) stained with ARG51752 anti-Histone H2A.X phospho (Ser139) antibody.
-
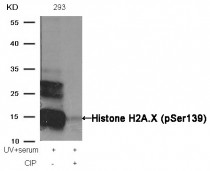
ARG51752 anti-Histone H2A.X phospho (Ser139) antibody WB image
Western blot: Extracts from 293 cells, treated with UV+serum or calf intestinal phosphatase (CIP), stained with ARG51752 anti-Histone H2A.X phospho (Ser139) antibody.
-
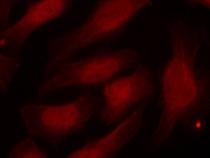
ARG51752 anti-Histone H2A.X phospho (Ser139) antibody ICC/IF image
Immunofluorescence: methanol-fixed HeLa cells stained with ARG51752 anti-Histone H2A.X phospho (Ser139) antibody.