ARG54153
anti-hHR23b antibody
anti-hHR23b antibody for ICC/IF,IHC-Formalin-fixed paraffin-embedded sections,Western blot and Human,Mouse,Rat,Hamster,Monkey
Gene Regulation antibody
Overview
| Product Description | Mouse Monoclonal antibody recognizes HR23B |
|---|---|
| Tested Reactivity | Hu, Ms, Rat, Hm, Mk |
| Tested Application | ICC/IF, IHC-P, WB |
| Host | Mouse |
| Clonality | Monoclonal |
| Isotype | IgG2b |
| Target Name | hHR23b |
| Antigen Species | Human |
| Immunogen | Purified recombinant human hHR23b protein fragments expressed in E.coli. |
| Conjugation | Un-conjugated |
| Alternate Names | HR23B; HHR23B; UV excision repair protein RAD23 homolog B; P58; hHR23B; p58; XP-C repair-complementing complex 58 kDa protein |
Application Instructions
| Application Suggestion |
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Application Note | IHC-P: Antigen Retrieval: Pressure cooking was performed in Citrate buffer (pH 6.0). * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. |
||||||||
| Observed Size | 58 kDa |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purified |
| Buffer | PBS (pH 7.4), 0.02% Sodium azide and 50% Glycerol |
| Preservative | 0.02% Sodium azide |
| Stabilizer | 50% Glycerol |
| Concentration | 1.5 mg/ml |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links | |
|---|---|
| Gene Symbol | RAD23B |
| Gene Full Name | RAD23 homolog B (S. cerevisiae) |
| Background | Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. May play a role in endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome._x000D_ Involved in global genome nucleotide excision repair (GG-NER) by acting as component of the XPC complex. Cooperatively with CETN2 appears to stabilize XPC. May protect XPC from proteasomal degradation._x000D_ The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. |
| Function | Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. May play a role in endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome. Involved in global genome nucleotide excision repair (GG-NER) by acting as component of the XPC complex. Cooperatively with CETN2 appears to stabilize XPC. May protect XPC from proteasomal degradation. The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. [UniProt] |
| Cellular Localization | Nucleus. Cytoplasm. Note: The intracellular distribution is cell cycle dependent. Localized to the nucleus and the cytoplasm during G1 phase. Nuclear levels decrease during S-phase; upon entering mitosis, relocalizes in the cytoplasm without association with chromatin. |
| Research Area | Gene Regulation antibody |
| Calculated MW | 43 kDa |
Images (3) Click the Picture to Zoom In
-
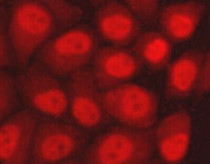
ARG54153 anti-hHR23b antibody ICC/IF image
Immunofluorescence: HeLa cells fixed with 4% Paraformaldehyde and stained with ARG54153 anti-hHR23b antibody at 1:100 dilution.
-
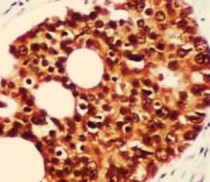
ARG54153 anti-hHR23b antibody IHC-P image
Immunohistochemistry: Paraffin-embedded Human prostate cancer stained with ARG54153 anti-hHR23b antibody at 1:100 dilution. Antigen Retrieval: Pressure cooking was performed in Citrate buffer (pH 6.0).
-
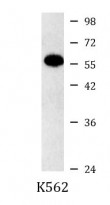
ARG54153 anti-hHR23b antibody WB image
Western blot: K562 cell lysate stained with ARG54153 anti-hHR23b antibody at 1:1000 dilution.