ARG56404
anti-XPC antibody
anti-XPC antibody for IHC-Formalin-fixed paraffin-embedded sections,Immunoprecipitation,Western blot and Human
Overview
| Product Description | Rabbit Polyclonal antibody recognizes XPC |
|---|---|
| Tested Reactivity | Hu |
| Tested Application | IHC-P, IP, WB |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Target Name | XPC |
| Antigen Species | Human |
| Immunogen | Recombinant protein of Human XPC |
| Conjugation | Un-conjugated |
| Alternate Names | p125; XPCC; Xeroderma pigmentosum group C-complementing protein; RAD4; XP3; DNA repair protein complementing XP-C cells |
Application Instructions
| Application Suggestion |
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. | ||||||||
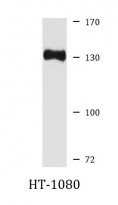
| Positive Control | HT-1080 |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purification with immunogen. |
| Buffer | PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. |
| Preservative | 0.02% sodium azide |
| Stabilizer | 50% Glycerol |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links |
Swiss-port # Q01831 Human DNA repair protein complementing XP-C cells |
|---|---|
| Gene Symbol | XPC |
| Gene Full Name | xeroderma pigmentosum, complementation group C |
| Background | This gene encodes a component of the nucleotide excision repair (NER) pathway. There are multiple components involved in the NER pathway, including Xeroderma pigmentosum (XP) A-G and V, Cockayne syndrome (CS) A and B, and trichothiodystrophy (TTD) group A, etc. This component, XPC, plays an important role in the early steps of global genome NER, especially in damage recognition, open complex formation, and repair protein complex formation. Mutations in this gene or some other NER components result in Xeroderma pigmentosum, a rare autosomal recessive disorder characterized by increased sensitivity to sunlight with the development of carcinomas at an early age. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Mar 2009] |
| Function | Involved in global genome nucleotide excision repair (GG-NER) by acting as damage sensing and DNA-binding factor component of the XPC complex. Has only a low DNA repair activity by itself which is stimulated by RAD23B and RAD23A. Has a preference to bind DNA containing a short single-stranded segment but not to damaged oligonucleotides. This feature is proposed to be related to a dynamic sensor function: XPC can rapidly screen duplex DNA for non-hydrogen-bonded bases by forming a transient nucleoprotein intermediate complex which matures into a stable recognition complex through an intrinsic single-stranded DNA-binding activity. The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. [UniProt] |
| Calculated MW | 106 kDa |
| PTM | Ubiquitinated upon UV irradiation; the ubiquitination requires the UV-DDB complex, appears to be reversible and does not serve as a signal for degradation (PubMed:15882621, PubMed:23751493). Ubiquitinated by RNF11 via 'Lys-63'-linked ubiquitination (PubMed:23751493). Ubiquitination by RNF111 is polysumoylation-dependent and promotes nucleotide excision repair (PubMed:23751493). Sumoylated; sumoylation promotes ubiquitination by RNF111. |
Images (1) Click the Picture to Zoom In