ARG59351
anti-SIRT2 antibody
anti-SIRT2 antibody for Western blot and Mouse,Rat
Overview
| Product Description | Mouse Monoclonal antibody recognizes SIRT2 |
|---|---|
| Tested Reactivity | Ms, Rat |
| Predict Reactivity | Hu |
| Tested Application | WB |
| Host | Mouse |
| Clonality | Monoclonal |
| Isotype | IgG2b |
| Target Name | SIRT2 |
| Antigen Species | Human |
| Immunogen | Purified Recombinant Human SIRT2 protein. |
| Conjugation | Un-conjugated |
| Alternate Names | SIR2-like protein 2; SIR2L2; NAD-dependent protein deacetylase sirtuin-2; EC 3.5.1.-; SIR2; SIR2L; Regulatory protein SIR2 homolog 2 |
Application Instructions
| Application Suggestion |
|
||||
|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purified. |
| Buffer | PBS (pH 7.4), 0.03% Proclin300 and 50% Glycerol. |
| Preservative | 0.03% Proclin300 |
| Stabilizer | 50% Glycerol |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links |
Swiss-port # Q5RJQ4 Rat NAD-dependent protein deacetylase sirtuin-2 Swiss-port # Q8VDQ8 Mouse NAD-dependent protein deacetylase sirtuin-2 |
|---|---|
| Gene Symbol | SIRT2 |
| Gene Full Name | sirtuin 2 |
| Background | This gene encodes a member of the sirtuin family of proteins, homologs to the yeast Sir2 protein. Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes. The functions of human sirtuins have not yet been determined; however, yeast sirtuin proteins are known to regulate epigenetic gene silencing and suppress recombination of rDNA. Studies suggest that the human sirtuins may function as intracellular regulatory proteins with mono-ADP-ribosyltransferase activity. The protein encoded by this gene is included in class I of the sirtuin family. Several transcript variants are resulted from alternative splicing of this gene. [provided by RefSeq, Jul 2010] |
| Function | NAD-dependent protein deacetylase, which deacetylates internal lysines on histone and alpha-tubulin as well as many other proteins such as key transcription factors. Participates in the modulation of multiple and diverse biological processes such as cell cycle control, genomic integrity, microtubule dynamics, cell differentiation, metabolic networks, and autophagy. Plays a major role in the control of cell cycle progression and genomic stability. Functions in the antephase checkpoint preventing precocious mitotic entry in response to microtubule stress agents, and hence allowing proper inheritance of chromosomes. Positively regulates the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase complex activity by deacetylating CDC20 and FZR1, then allowing progression through mitosis. Associates both with chromatin at transcriptional start sites (TSSs) and enhancers of active genes. Plays a role in cell cycle and chromatin compaction through epigenetic modulation of the regulation of histone H4 'Lys-20' methylation (H4K20me1) during early mitosis. Specifically deacetylates histone H4 at 'Lys-16' (H4K16ac) between the G2/M transition and metaphase enabling H4K20me1 deposition by SETD8 leading to ulterior levels of H4K20me2 and H4K20me3 deposition throughout cell cycle, and mitotic S-phase progression. Deacetylates SETD8 modulating SETD8 chromatin localization during the mitotic stress response. Deacetylates also histone H3 at 'Lys-57' (H3K56ac) during the mitotic G2/M transition. Upon bacterium Listeria monocytogenes infection, deacetylates 'Lys-18' of histone H3 in a receptor tyrosine kinase MET- and PI3K/Akt-dependent manner, thereby inhibiting transcriptional activity and promoting late stages of listeria infection. During oocyte meiosis progression, may deacetylate histone H4 at 'Lys-16' (H4K16ac) and alpha-tubulin, regulating spindle assembly and chromosome alignment by influencing microtubule dynamics and kinetochore function. Deacetylates alpha-tubulin at 'Lys-40' and hence controls neuronal motility, oligodendroglial cell arbor projection processes and proliferation of non-neuronal cells. Phosphorylation at Ser-368 by a G1/S-specific cyclin E-CDK2 complex inactivates SIRT2-mediated alpha-tubulin deacetylation, negatively regulating cell adhesion, cell migration and neurite outgrowth during neuronal differentiation. Deacetylates PARD3 and participates in the regulation of Schwann cell peripheral myelination formation during early postnatal development and during postinjury remyelination. Involved in several cellular metabolic pathways. Plays a role in the regulation of blood glucose homeostasis by deacetylating and stabilizing phosphoenolpyruvate carboxykinase PCK1 activity in response to low nutrient availability. Acts as a key regulator in the pentose phosphate pathway (PPP) by deacetylating and activating the glucose-6-phosphate G6PD enzyme, and therefore, stimulates the production of cytosolic NADPH to counteract oxidative damage. Maintains energy homeostasis in response to nutrient deprivation as well as energy expenditure by inhibiting adipogenesis and promoting lipolysis. Attenuates adipocyte differentiation by deacetylating and promoting FOXO1 interaction to PPARG and subsequent repression of PPARG-dependent transcriptional activity. Plays a role in the regulation of lysosome-mediated degradation of protein aggregates by autophagy in neuronal cells. Deacetylates FOXO1 in response to oxidative stress or serum deprivation, thereby negatively regulating FOXO1-mediated autophagy. Deacetylates a broad range of transcription factors and co-regulators regulating target gene expression. Deacetylates transcriptional factor FOXO3 stimulating the ubiquitin ligase SCF(SKP2)-mediated FOXO3 ubiquitination and degradation. Deacetylates HIF1A and therefore promotes HIF1A degradation and inhibition of HIF1A transcriptional activity in tumor cells in response to hypoxia. Deacetylates RELA in the cytoplasm inhibiting NF-kappaB-dependent transcription activation upon TNF-alpha stimulation. Inhibits transcriptional activation by deacetylating p53/TP53 and EP300. Deacetylates also EIF5A. Functions as a negative regulator on oxidative stress-tolerance in response to anoxia-reoxygenation conditions. Plays a role as tumor suppressor. Isoform 1: Deacetylates EP300, alpha-tubulin and histone H3 and H4. Isoform 2: Deacetylates EP300, alpha-tubulin and histone H3 and H4. Isoform 5: Lacks deacetylation activity. [UniProt] |
| Calculated MW | 43 kDa |
| PTM | Phosphorylated at phosphoserine and phosphothreonine. Phosphorylated at Ser-368 by a mitotic kinase CDK1/cyclin B at the G2/M transition; phosphorylation regulates the delay in cell-cycle progression. Phosphorylated at Ser-368 by a mitotic kinase G1/S-specific cyclin E/Cdk2 complex; phosphorylation inactivates SIRT2-mediated alpha-tubulin deacetylation and thereby negatively regulates cell adhesion, cell migration and neurite outgrowth during neuronal differentiation. Phosphorylated by cyclin A/Cdk2 and p35-Cdk5 complexes and to a lesser extent by the cyclin D3/Cdk4 and cyclin B/Cdk1, in vitro. Dephosphorylated at Ser-368 by CDC14A and CDC14B around early anaphase. Acetylated by EP300; acetylation leads both to the decreased of SIRT2-mediated alpha-tubulin deacetylase activity and SIRT2-mediated down-regulation of TP53 transcriptional activity. Ubiquitinated. [UniProt] |
Images (2) Click the Picture to Zoom In
-
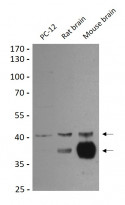
ARG59351 anti-SIRT2 antibody WB image
Western blot: PC-12, Rat brain and Mouse brain lysates stained with ARG59351 anti-SIRT2 antibody at 1:1000 dilution.
-
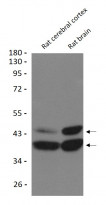
ARG59351 anti-SIRT2 antibody WB image
Western blot: Rat cerebral cortex and Rat brain lysates stained with ARG59351 anti-SIRT2 antibody at 1:1000 dilution.