ARG54762
anti-Histone H3 monomethyl (Lys9) antibody - ChIP-Grade
anti-Histone H3 monomethyl (Lys9) antibody - ChIP-Grade for ChIP,ChIP-sequencing,ICC/IF,IHC-Formalin-fixed paraffin-embedded sections,Western blot and Human,Mouse,Rat
Gene Regulation antibody
Overview
| Product Description | Rabbit Polyclonal antibody recognizes Histone H3 monomethyl (Lys9) - ChIP-Grade |
|---|---|
| Tested Reactivity | Hu, Ms, Rat |
| Tested Application | ChIP, ChIP-seq, ICC/IF, IHC-P, WB |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Target Name | Histone H3 |
| Antigen Species | Human |
| Immunogen | Synthetic methylated peptide around Lys9 of Human histone H3 (NP_003484.1) |
| Conjugation | Un-conjugated |
| Alternate Names | H3FT; H3/g; Histone H3.1t; H3t; H3/t; H3.4 |
Application Instructions
| Application Suggestion |
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. | ||||||||||||
| Positive Control | HeLa |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purification with immunogen. |
| Buffer | PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol |
| Preservative | 0.02% Sodium azide |
| Stabilizer | 50% Glycerol |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links | |
|---|---|
| Gene Symbol | HIST3H3 |
| Gene Full Name | histone cluster 3, H3 |
| Background | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. Histone H3 gene is intronless and encodes a replication-dependent histone that is a member of the histone H3 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is located separately from the other H3 genes that are in the histone gene cluster on chromosome 6p22-p21.3. [provided by RefSeq, Aug 2015] |
| Function | Histone H3 is a core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. [UniProt] |
| Research Area | Gene Regulation antibody |
| Calculated MW | 16 kDa |
| PTM | Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability (By similarity). Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription. Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters (By similarity). Methylation at Lys-5 (H3K4me), Lys-37 (H3K36me) and Lys-80 (H3K79me) are linked to gene activation. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are linked to gene repression. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication (By similarity). Phosphorylated at Thr-4 (H3T3ph) by GSG2/haspin during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin (By similarity). Ubiquitinated. Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression (By similarity). |
Images (3) Click the Picture to Zoom In
-
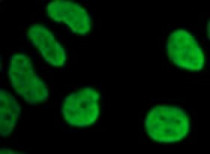
ARG54762 anti-Histone H3 monomethyl (Lys9) antibody - ChIP-Grade ICC/IF image
Immunofluorescence: 293T cells stained with ARG54762 anti-Histone H3 monomethyl (Lys9) antibody (ChIP-Grade).
-
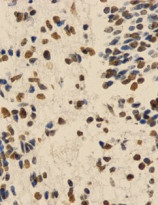
ARG54762 anti-Histone H3 monomethyl (Lys9) antibody - ChIP-Grade IHC-P image
Immunohistochemistry: Paraffin-embedded Human embryo brain tissue stained with ARG54762 anti-Histone H3 monomethyl (Lys9) antibody (ChIP-Grade) at 1:200 dilution.
-
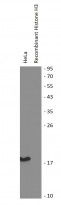
ARG54762 anti-Histone H3 monomethyl (Lys9) antibody - ChIP-Grade WB image
Western blot: HeLa cell lysate and Recombinant Histone H3 protein expressed in E. coli (negative control) stained with ARG54762 anti-Histone H3 monomethyl (Lys9) antibody (ChIP-Grade).