ARG55293
anti-APE1 antibody
anti-APE1 antibody for ICC/IF,IHC-Formalin-fixed paraffin-embedded sections,Immunoprecipitation,Western blot and Human
Cell Biology and Cellular Response antibody; Gene Regulation antibody

Overview
| Product Description | Rabbit Polyclonal antibody recognizes APE1 |
|---|---|
| Tested Reactivity | Hu |
| Tested Application | ICC/IF, IHC-P, IP, WB |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Target Name | APE1 |
| Antigen Species | Human |
| Immunogen | Recombinant protein of Human APE1 |
| Conjugation | Un-conjugated |
| Alternate Names | APE; APX; APE1; APEN; APEX; HAP1; REF1; DNA-(apurinic or apyrimidinic site) lyase; EC 3.1.-.-; EC 4.2.99.18; APEX nuclease; APEN; Apurinic-apyrimidinic endonuclease 1; AP endonuclease 1; APE-1; REF-1; Redox factor-1 |
Application Instructions
| Application Suggestion |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. | ||||||||||
| Positive Control | HepG2 and 293T |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purification with immunogen. |
| Buffer | PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol |
| Preservative | 0.02% Sodium azide |
| Stabilizer | 50% Glycerol |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links |
Swiss-port # P27695 Human DNA-(apurinic or apyrimidinic site) lyase |
|---|---|
| Gene Symbol | APEX1 |
| Gene Full Name | APEX nuclease (multifunctional DNA repair enzyme) 1 |
| Background | Apurinic/apyrimidinic (AP) sites occur frequently in DNA molecules by spontaneous hydrolysis, by DNA damaging agents or by DNA glycosylases that remove specific abnormal bases. AP sites are pre-mutagenic lesions that can prevent normal DNA replication so the cell contains systems to identify and repair such sites. Class II AP endonucleases cleave the phosphodiester backbone 5' to the AP site. This gene encodes the major AP endonuclease in human cells. Splice variants have been found for this gene; all encode the same protein. [provided by RefSeq, Jul 2008] |
| Function | Multifunctional protein that plays a central role in the cellular response to oxidative stress. The two major activities of APEX1 in DNA repair and redox regulation of transcriptional factors. Functions as a apurinic/apyrimidinic (AP) endodeoxyribonuclease in the DNA base excision repair (BER) pathway of DNA lesions induced by oxidative and alkylating agents. Initiates repair of AP sites in DNA by catalyzing hydrolytic incision of the phosphodiester backbone immediately adjacent to the damage, generating a single-strand break with 5'-deoxyribose phosphate and 3'-hydroxyl ends. Does also incise at AP sites in the DNA strand of DNA/RNA hybrids, single-stranded DNA regions of R-loop structures, and single-stranded RNA molecules. Has a 3'-5' exoribonuclease activity on mismatched deoxyribonucleotides at the 3' termini of nicked or gapped DNA molecules during short-patch BER. Possesses a DNA 3' phosphodiesterase activity capable of removing lesions (such as phosphoglycolate) blocking the 3' side of DNA strand breaks. May also play a role in the epigenetic regulation of gene expression by participating in DNA demethylation. Acts as a loading factor for POLB onto non-incised AP sites in DNA and stimulates the 5'-terminal deoxyribose 5'-phosphate (dRp) excision activity of POLB. Plays a role in the protection from granzymes-mediated cellular repair leading to cell death. Also involved in the DNA cleavage step of class switch recombination (CSR). On the other hand, APEX1 also exerts reversible nuclear redox activity to regulate DNA binding affinity and transcriptional activity of transcriptional factors by controlling the redox status of their DNA-binding domain, such as the FOS/JUN AP-1 complex after exposure to IR. Involved in calcium-dependent down-regulation of parathyroid hormone (PTH) expression by binding to negative calcium response elements (nCaREs). Together with HNRNPL or the dimer XRCC5/XRCC6, associates with nCaRE, acting as an activator of transcriptional repression. Stimulates the YBX1-mediated MDR1 promoter activity, when acetylated at Lys-6 and Lys-7, leading to drug resistance. Acts also as an endoribonuclease involved in the control of single-stranded RNA metabolism. Plays a role in regulating MYC mRNA turnover by preferentially cleaving in between UA and CA dinucleotides of the MYC coding region determinant (CRD). In association with NMD1, plays a role in the rRNA quality control process during cell cycle progression. Associates, together with YBX1, on the MDR1 promoter. Together with NPM1, associates with rRNA. Binds DNA and RNA. [UniProt] |
| Research Area | Cell Biology and Cellular Response antibody; Gene Regulation antibody |
| Calculated MW | 36 kDa |
| PTM | Phosphorylated. Phosphorylation by kinase PKC or casein kinase CK2 results in enhanced redox activity that stimulates binding of the FOS/JUN AP-1 complex to its cognate binding site. AP-endodeoxyribonuclease activity is not affected by CK2-mediated phosphorylation. Phosphorylation of Thr-233 by CDK5 reduces AP-endodeoxyribonuclease activity resulting in accumulation of DNA damage and contributing to neuronal death. Acetylated on Lys-6 and Lys-7. Acetylation is increased by the transcriptional coactivator EP300 acetyltransferase, genotoxic agents like H(2)O(2) and methyl methanesulfonate (MMS). Acetylation increases its binding affinity to the negative calcium response element (nCaRE) DNA promoter. The acetylated form induces a stronger binding of YBX1 to the Y-box sequence in the MDR1 promoter than the unacetylated form. Deacetylated on lysines. Lys-6 and Lys-7 are deacetylated by SIRT1. Cleaved at Lys-31 by granzyme A to create the mitochondrial form; leading in reduction of binding to DNA, AP endodeoxynuclease activity, redox activation of transcription factors and to enhanced cell death. Cleaved by granzyme K; leading to intracellular ROS accumulation and enhanced cell death after oxidative stress. Cys-65 and Cys-93 are nitrosylated in response to nitric oxide (NO) and lead to the exposure of the nuclear export signal (NES). Ubiquitinated by MDM2; leading to translocation to the cytoplasm and proteasomal degradation. |
Images (4) Click the Picture to Zoom In
-
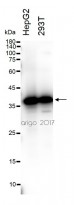
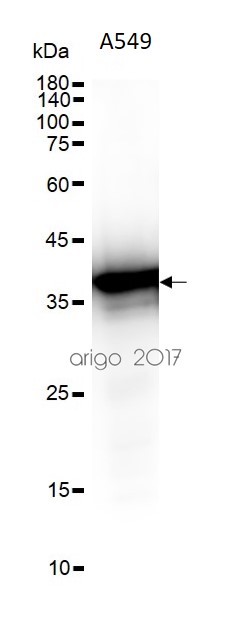
ARG55293 anti-APE1 antibody WB image
Western blot: 30 µg of HepG2 and 293T cell lysates stained with ARG55293 anti-APE1 antibody at 1:500 dilution.
-
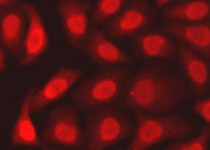
ARG55293 anti-APE1 antibody ICC/IF image
Immunofluorescence: A549 cells stained with ARG55293 anti-APE1 antibody.
-
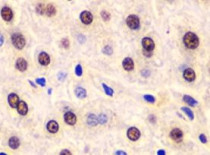
ARG55293 anti-APE1 antibody IHC-P image
Immunohistochemistry: Paraffin-embedded Human liver injury stained with ARG55293 anti-APE1 antibody at 1:100 dilution.
-
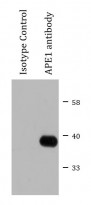
ARG55293 anti-APE1 antibody IP image
Immunoprecipitation: 200 µg extracts of HeLa cells immunoprecipitated and stained with ARG55293 anti-APE1 antibody at 1:1000 dilution.